RBP–lncRNA interactions were available at http://starbase.sysu.edu.cn/rbpLncRNA.php.
an invited paper described these protein–lncRNA interactions has been published in Frontiers journal.
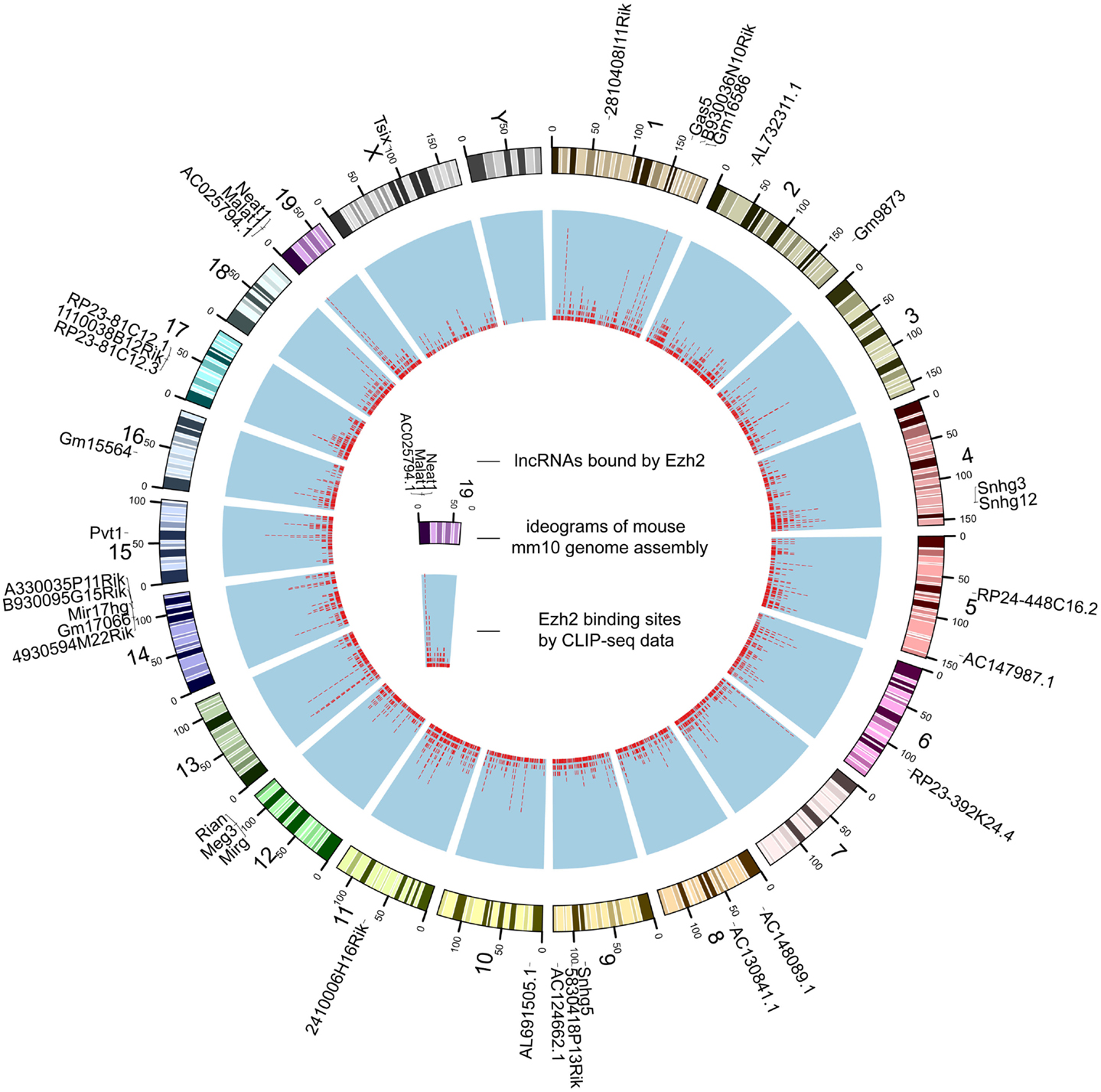
The genome-wide binding map of Ezh2 in mouse.
You are currently viewing the SEQanswers forums as a guest, which limits your access. Click here to register now, and join the discussion






| Topics | Statistics | Last Post | ||
|---|---|---|---|---|
|
Started by seqadmin, 04-11-2024, 12:08 PM
|
0 responses
22 views
0 likes
|
Last Post
by seqadmin
04-11-2024, 12:08 PM
|
||
|
Started by seqadmin, 04-10-2024, 10:19 PM
|
0 responses
24 views
0 likes
|
Last Post
by seqadmin
04-10-2024, 10:19 PM
|
||
|
Started by seqadmin, 04-10-2024, 09:21 AM
|
0 responses
19 views
0 likes
|
Last Post
by seqadmin
04-10-2024, 09:21 AM
|
||
|
Started by seqadmin, 04-04-2024, 09:00 AM
|
0 responses
50 views
0 likes
|
Last Post
by seqadmin
04-04-2024, 09:00 AM
|
Leave a comment: