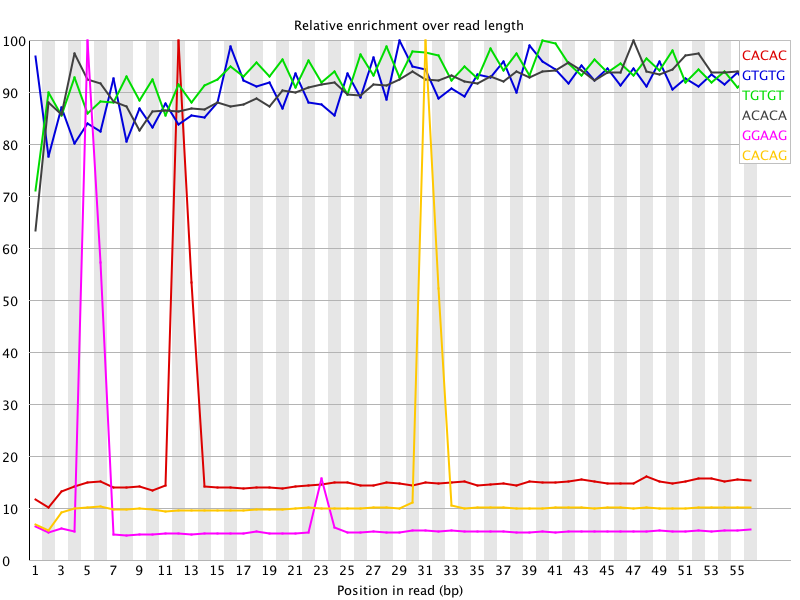
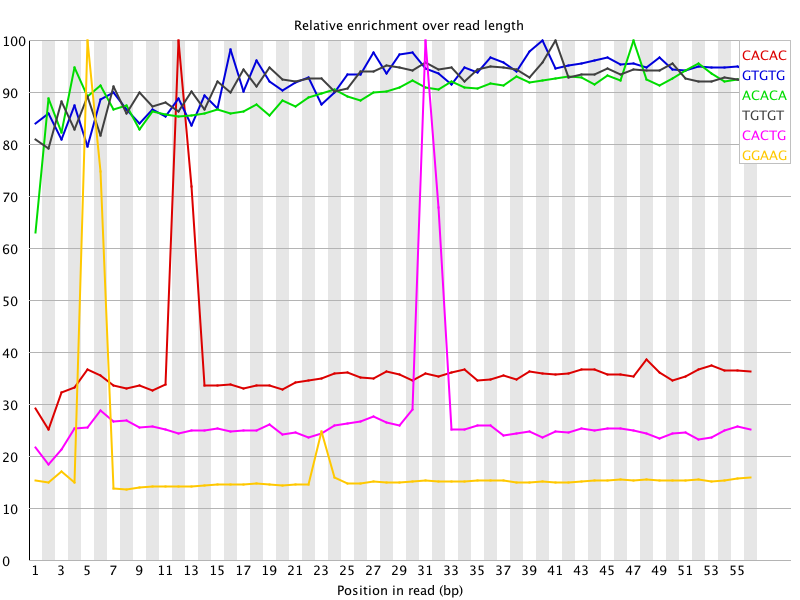
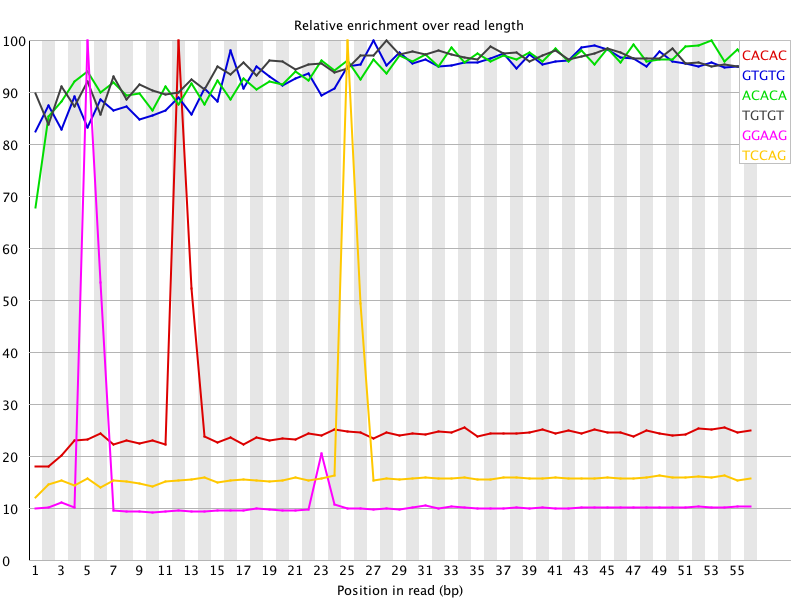
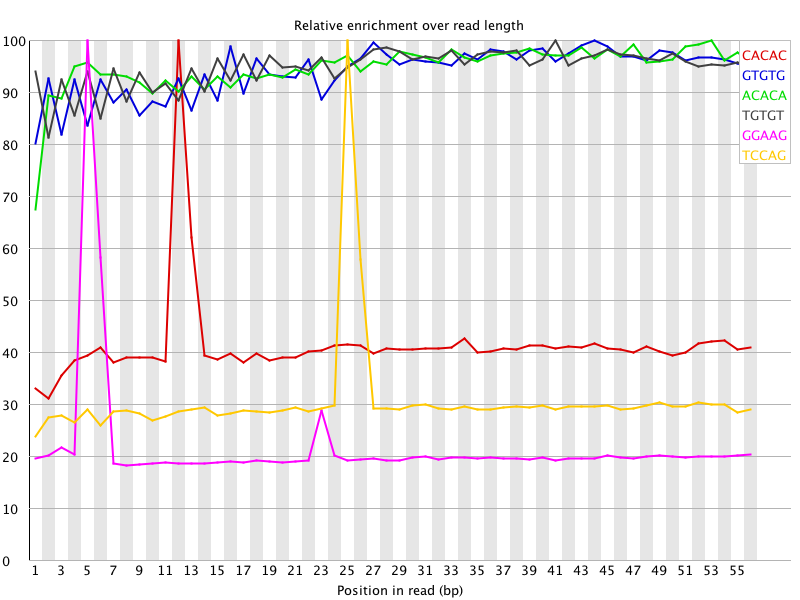
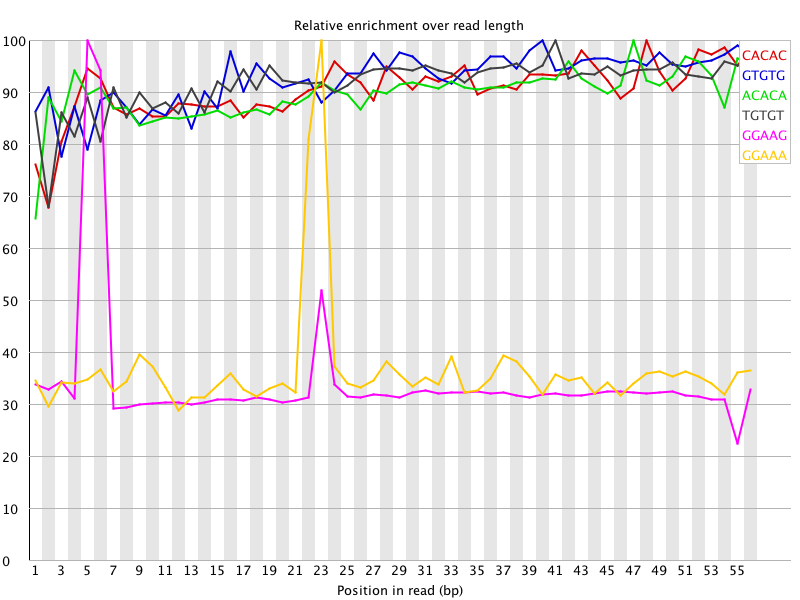
I ran FatsQC on a set of 4 Illumina PE exomes and got really weird graphs for the kmer distribution. I've never seen this pattern before - does anyone know what is going on and what to do about it?




You are currently viewing the SEQanswers forums as a guest, which limits your access. Click here to register now, and join the discussion
| Topics | Statistics | Last Post | ||
|---|---|---|---|---|
|
Started by seqadmin, Today, 11:09 AM
|
0 responses
22 views
0 likes
|
Last Post
by seqadmin
Today, 11:09 AM
|
||
|
Started by seqadmin, Today, 06:13 AM
|
0 responses
20 views
0 likes
|
Last Post
by seqadmin
Today, 06:13 AM
|
||
|
Started by seqadmin, 11-01-2024, 06:09 AM
|
0 responses
30 views
0 likes
|
Last Post
by seqadmin
11-01-2024, 06:09 AM
|
||
|
New Model Aims to Explain Polygenic Diseases by Connecting Genomic Mutations and Regulatory Networks
by seqadmin
Started by seqadmin, 10-30-2024, 05:31 AM
|
0 responses
21 views
0 likes
|
Last Post
by seqadmin
10-30-2024, 05:31 AM
|
Comment