Seqanswers Leaderboard Ad
Collapse
Announcement
Collapse
No announcement yet.
X
-
thank you very much again, someone told me that the dataset can be fitting with R package "spline.smooth" first and then make ANCOVA ~
-
Hmmm good question. I think someone with a stats background (i.e. not me) could answer your question accurately, but I'll have a guess:
I think you would do two separate ANCOVAs, one each for you two age brackets. From these I believe you could compare the correlation p values, but not the slopes i.e. the effect size. For this I think you would need to use something like as Tukey's ad hoc test.
Don't hold me to that!
You should get advice from someone with more knowledge on the matter than I can provide.
J
Leave a comment:
-
thank you for your information. and here i want to ask how to handle the dataset. because the age and number of samples are different. the data is not paired. like ancova(x,y), the vector x,y should be same length and paired, right?
Leave a comment:
-
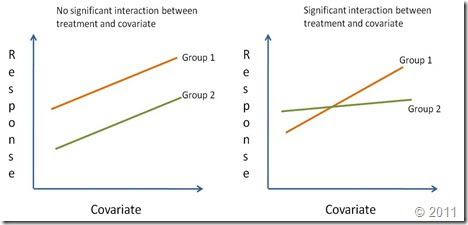
how to make ANCOVA in two indenpendent dataset
for example, there are two gene expression microarray datasets,
samples 1
gene 14 year-old 25 year-old .....
LDH 0.2 0.8 ....
samples 2
gene 12 year-old 20year-old .....
LDH 0.25 0.78
if i know the LDH gene expression level is increasing with age, I want to know the correlataion of the gene expression change in aging process between two samples, how i calculated the Pearson correlation. and how I make a ANCOVA in the above LDH expression between the two samples.Tags: None
Latest Articles
Collapse
-
by seqadmin
The complexity of cancer is clearly demonstrated in the diverse ecosystem of the tumor microenvironment (TME). The TME is made up of numerous cell types and its development begins with the changes that happen during oncogenesis. “Genomic mutations, copy number changes, epigenetic alterations, and alternative gene expression occur to varying degrees within the affected tumor cells,” explained Andrea O’Hara, Ph.D., Strategic Technical Specialist at Azenta. “As...-
Channel: Articles
07-08-2024, 03:19 PM -
ad_right_rmr
Collapse
News
Collapse
| Topics | Statistics | Last Post | ||
|---|---|---|---|---|
|
Started by seqadmin, 07-25-2024, 06:46 AM
|
0 responses
9 views
0 likes
|
Last Post
by seqadmin
07-25-2024, 06:46 AM
|
||
|
Started by seqadmin, 07-24-2024, 11:09 AM
|
0 responses
26 views
0 likes
|
Last Post
by seqadmin
07-24-2024, 11:09 AM
|
||
|
Started by seqadmin, 07-19-2024, 07:20 AM
|
0 responses
160 views
0 likes
|
Last Post
by seqadmin
07-19-2024, 07:20 AM
|
||
|
Started by seqadmin, 07-16-2024, 05:49 AM
|
0 responses
127 views
0 likes
|
Last Post
by seqadmin
07-16-2024, 05:49 AM
|

Leave a comment: