I'm currently looking for a way to find directed interactions in some specific pathways and visualizing them. I've been using Cytoscape up until now, but I'm a bit stuck on the directionality part (an other minor things I won't mention). I have my own interaction data, but without the directionality, which is the problem. I would either like to complement my own data with directionality between proteins, or start "fresh" with other interaction data that already includes directionality.
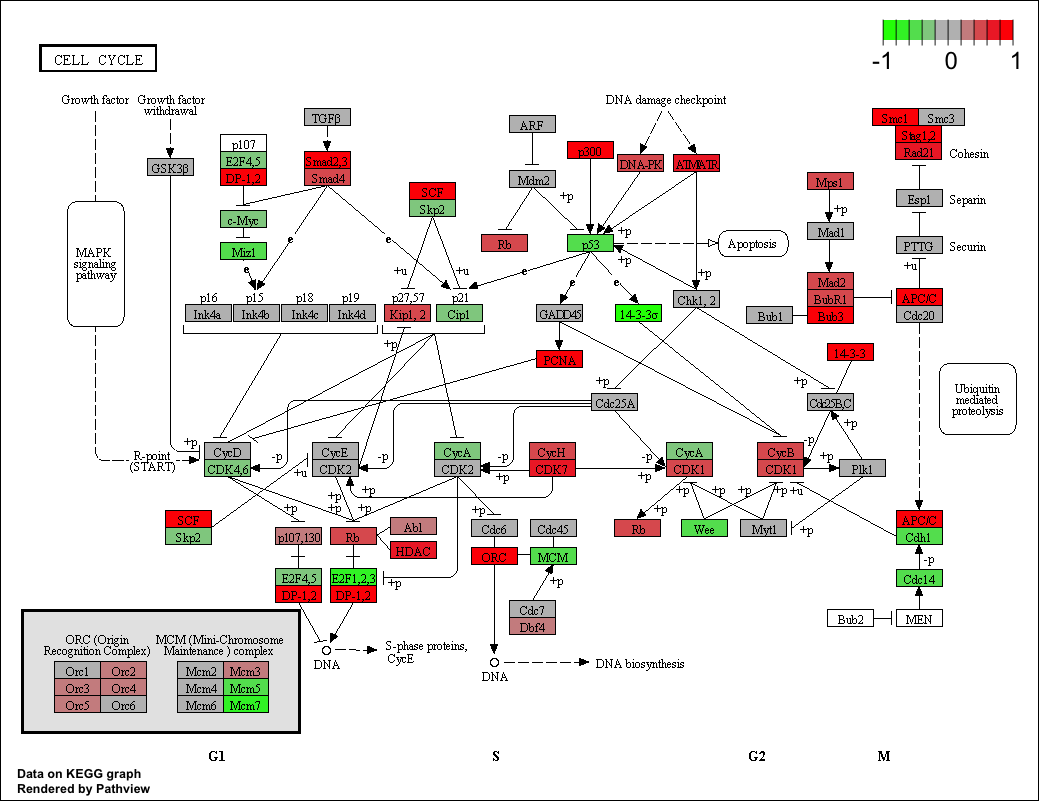
KEGG and Reactome seems to be two big interaction databases that at least seems to have directionality in them. I can, for example, look at the KEGG MAPK/ERK pathway and see that they have directionality in them. I can use the KEGG API to access the genes that constitute the pathway, but as far as I can tell I don't get the directionality from that.
I've also tried importing my genes directly into Cytoscape using the "public databases import" function (Reactome data), but there I am again confused as to how to get the directionality.
In essence, I just want a pathway map with directionality in Cytoscape (or other visualizing software) that I can use to visualize my own RNA-seq data (i.e. differential expression). Is this something that can be done, at all?
KEGG and Reactome seems to be two big interaction databases that at least seems to have directionality in them. I can, for example, look at the KEGG MAPK/ERK pathway and see that they have directionality in them. I can use the KEGG API to access the genes that constitute the pathway, but as far as I can tell I don't get the directionality from that.
I've also tried importing my genes directly into Cytoscape using the "public databases import" function (Reactome data), but there I am again confused as to how to get the directionality.
In essence, I just want a pathway map with directionality in Cytoscape (or other visualizing software) that I can use to visualize my own RNA-seq data (i.e. differential expression). Is this something that can be done, at all?


Comment