Hello,
Can someone help me understand my FastQC analysis?
The questions I am having are:
Do I need to cut my index primers?
Do per base sequence content and per base GC content graphs tell me that there is something wrong with my samples?
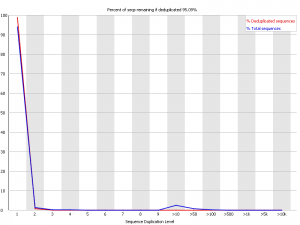
Also I don't understand what could be the cause of 10+ duplication level?!
Last thing I don't understand the Kmer graph. I watched a video that one could easily figure out the adapters used for the sequencing but my Kmer graph is so confusing I cannot understand anything!
Your help would be very much appreciated.
Parham
Can someone help me understand my FastQC analysis?
The questions I am having are:
Do I need to cut my index primers?
Do per base sequence content and per base GC content graphs tell me that there is something wrong with my samples?
Also I don't understand what could be the cause of 10+ duplication level?!
Last thing I don't understand the Kmer graph. I watched a video that one could easily figure out the adapters used for the sequencing but my Kmer graph is so confusing I cannot understand anything!
Your help would be very much appreciated.
Parham
Comment