Hi Guys,
So I'm trying to amplify whole mtDNA of a few species using long-range PCR. I've found a conserved region and designed long (30mer) primers at 3 different sites which are about 120 bp and 500 bp apart. When I tested my primers by amplifying the small region in-between the primers I get beautiful bright bands for each pair. But when I use the reverse compliments of the primers to try for the whole 15 kbp, I get a smear and a few non-specific bands. I'm stumped. Primers are long, priming site is conserved, PCR is a 2 step with high Ta. Don't know what else to try. Ideas? PCR conditions as follows:
94C - 1 min
98C - 10 sec
68C - 15 min
(Last 2 steps X 33 cycles)
72C - 10
PCR recipe (25ul reaction):
dH2O - 12.25ul
10X buffer - 2.5ul
dNTP - 4.0
Primer F - 1.0
Primer R - 1.0
Taq - 0.25ul
Template - 4.0 ul (2-4ng)
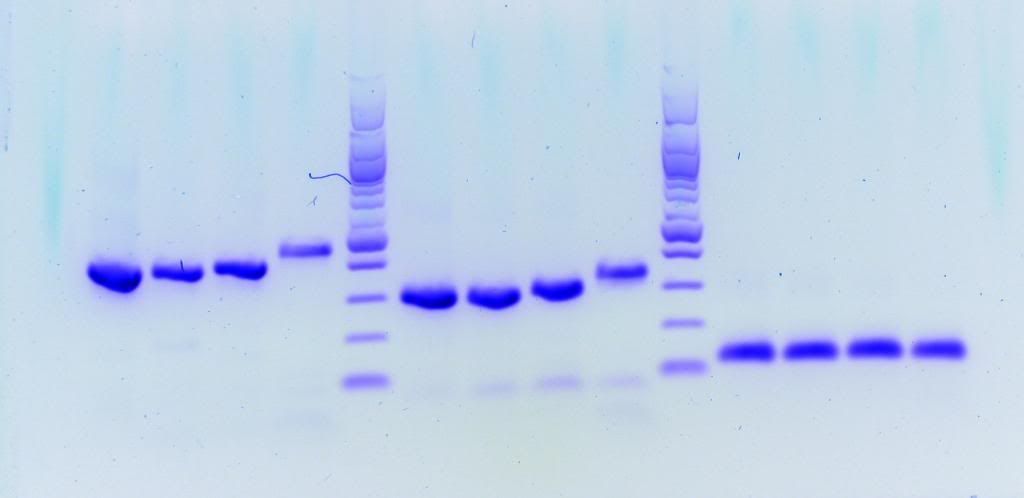
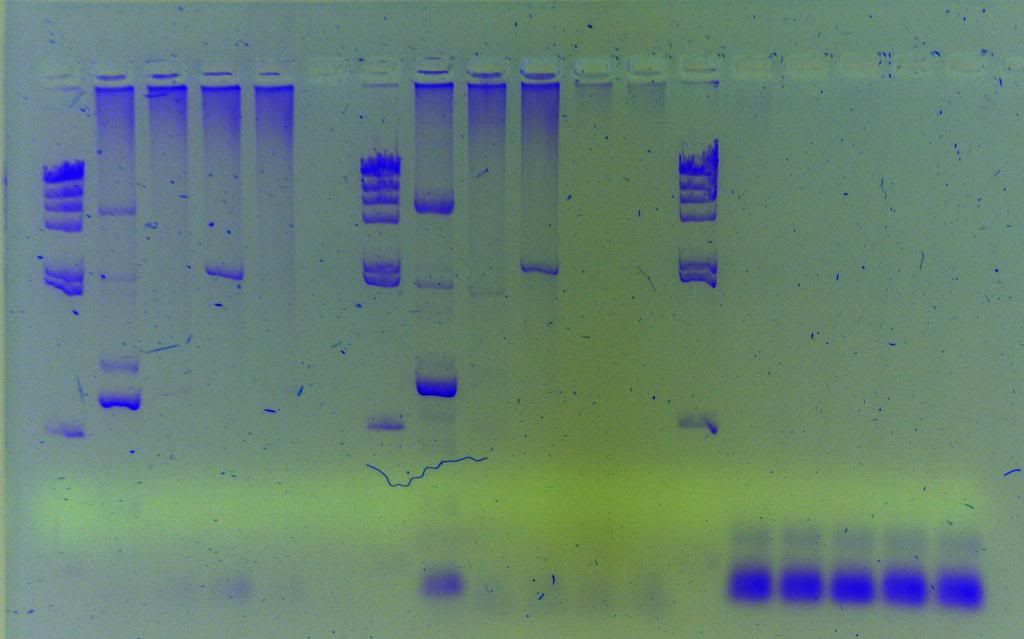
Taq is Takara LA which is a high end long-range enzyme supposedly capable of amplifying up to 40 kbp. First picture is small amplicons proving priming sites are good, second image is the results for my long-range. HindIII ladder in second image.
So I'm trying to amplify whole mtDNA of a few species using long-range PCR. I've found a conserved region and designed long (30mer) primers at 3 different sites which are about 120 bp and 500 bp apart. When I tested my primers by amplifying the small region in-between the primers I get beautiful bright bands for each pair. But when I use the reverse compliments of the primers to try for the whole 15 kbp, I get a smear and a few non-specific bands. I'm stumped. Primers are long, priming site is conserved, PCR is a 2 step with high Ta. Don't know what else to try. Ideas? PCR conditions as follows:
94C - 1 min
98C - 10 sec
68C - 15 min
(Last 2 steps X 33 cycles)
72C - 10
PCR recipe (25ul reaction):
dH2O - 12.25ul
10X buffer - 2.5ul
dNTP - 4.0
Primer F - 1.0
Primer R - 1.0
Taq - 0.25ul
Template - 4.0 ul (2-4ng)
Taq is Takara LA which is a high end long-range enzyme supposedly capable of amplifying up to 40 kbp. First picture is small amplicons proving priming sites are good, second image is the results for my long-range. HindIII ladder in second image.


Comment