Over christmas GRCh38, the newest human reference genome assembly, was released.Internally we have been using chromosome 20 of human reference builds to benchmark tools and pipelines with datasets. A BWA sequence alignment of the same dataset, generated on a HiSeq 2500, across the last major release GRCh37.69 and the new GRCh38 was performed.

Quantifiably, GRCh38 is very similar to the later GRCh37 releases, showing a change rate of 1 change every 159,558 bases on 37.69 and 1 change every 156,779 bases on 38 for our chromosome 20 dataset.Ts/Tv ratios between the two alignments of the same data across the two references to be quite similar at 0.3527 and 0.3445, respectively. Back of the envelope math seems to give a Δ of +19,359 between GRCh37.69 and 38.
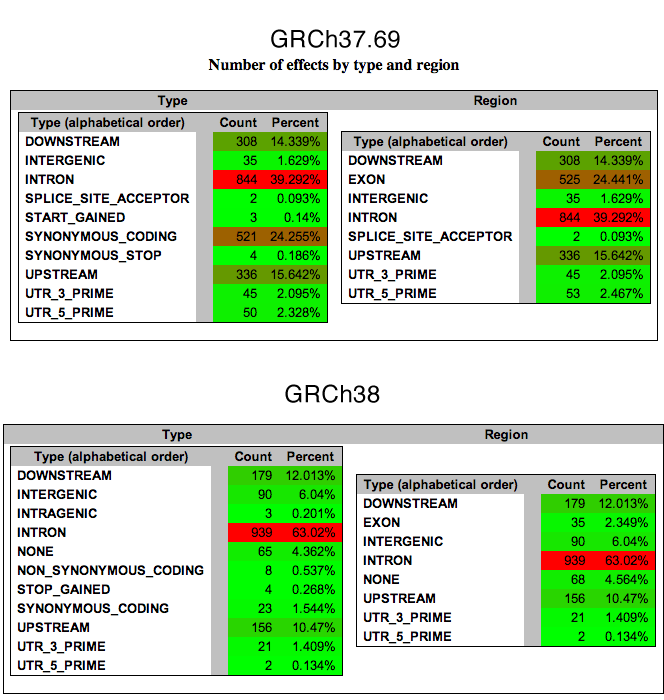
Annotations have a large deviation, to be expected for now.
Read the rest here: http://wp.me/pJHIj-nm

Quantifiably, GRCh38 is very similar to the later GRCh37 releases, showing a change rate of 1 change every 159,558 bases on 37.69 and 1 change every 156,779 bases on 38 for our chromosome 20 dataset.Ts/Tv ratios between the two alignments of the same data across the two references to be quite similar at 0.3527 and 0.3445, respectively. Back of the envelope math seems to give a Δ of +19,359 between GRCh37.69 and 38.
Annotations have a large deviation, to be expected for now.

Read the rest here: http://wp.me/pJHIj-nm
Comment